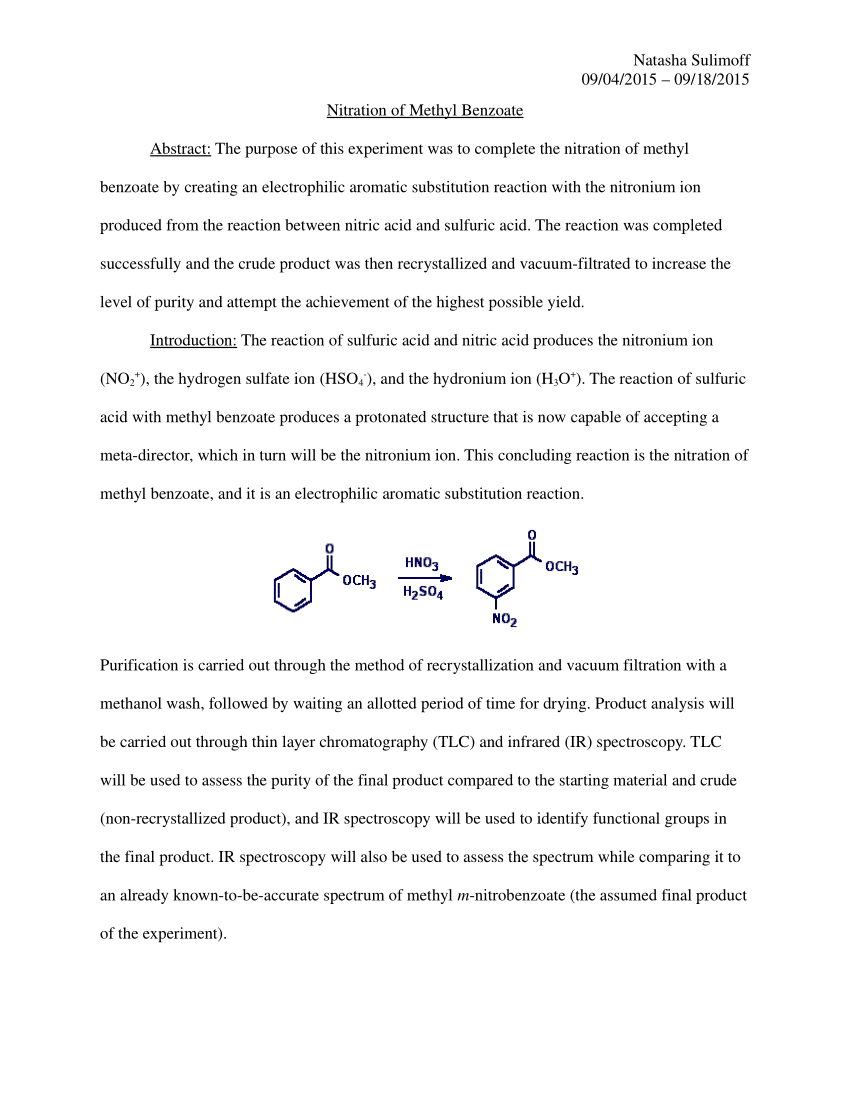
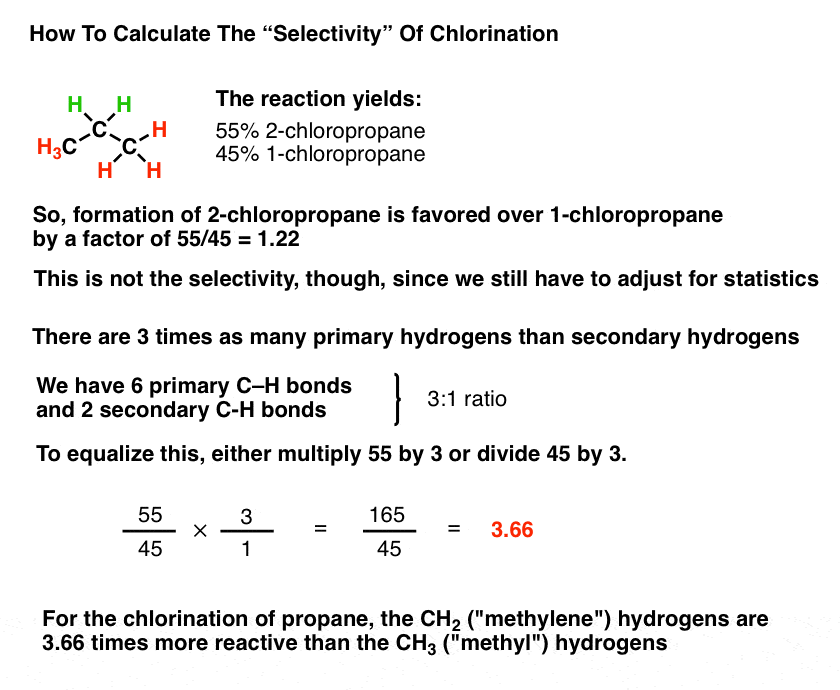
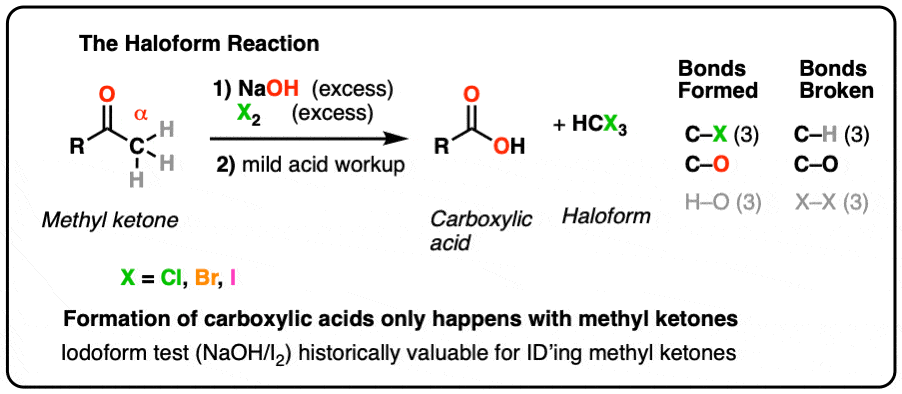
calculate ka for methyl red based on your results
How can bisulphite be used to measure DNA methylation levels?
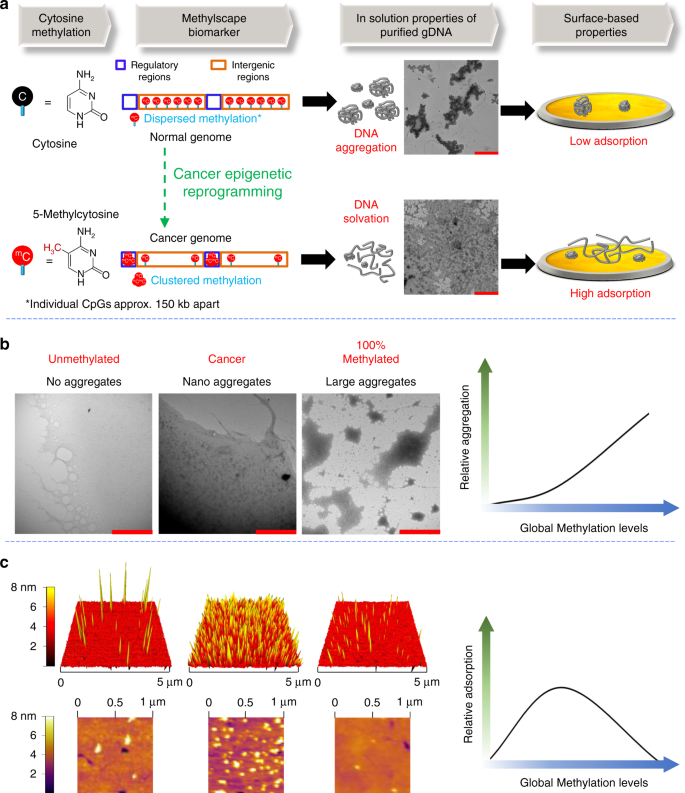
Bisulphite treatment in combination with specially designed genotyping microarrays makes it possible to measure DNA methylation levels at a preselected fraction of Cs throughout the genome. One key advantage of this approach is the (currently) lower per-sample cost compared with whole-genome bisulphite sequencing.
How is DNA methylation level calculated?
Reads with more than one perfect alignment with the reference sequence are discarded (greyed out), and for each CpG in the genomic DNA sequence, the DNA methylation level (bottom) is calculated as the percentage of aligning Cs among all uniquely mapped reads.
How is methylation calculated in qPCR?
To perform it, two pairs of primers are designed; one pair that favours amplification of methylated and another one of unmethylated DNA. Two qPCR reactions are performed for each sample, and relative methylation is calculated based on the difference of their Ct values.
How can a Bayesian network be used to determine DNA methylation levels?
Bayesian networks have been used for inferring absolute DNA methylation levels for single CpGs from the complex fragment distributions 62, and a bioinformatic analysis pipeline has been described for processing data obtained with the HELP–seq assay 63.
Description
The function calculates differential methylation statistics between two groups of samples. The function uses either logistic regression testor Fisher's Exact test to calculate differential methylation. See the rest of the help page and references for detailed explanation on statistics. rdrr.io
Value
a methylDiffobject containing the differential methylation statistics and locations for regions or bases rdrr.io
Details
Covariates can be included in the analysis. The function will then try to separate the influence of the covariates from the treatment effect via a linear model. The Chisq-test is used per default only when no overdispersion correction isapplied.If overdispersion correction is applied, the function automatically switches to the F-test. The Chisq-tes
References
Altuna Akalin, Matthias Kormaksson, Sheng Li,Francine E. Garrett-Bakelman, Maria E. Figueroa, Ari Melnick, Christopher E. Mason. (2012). "methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles." Genome Biology. McCullagh and Nelder. (1989). Generalized Linear Models. Chapmanand Hall. London New York. Barnard.
|
The pKa of methyl red will be determined by measuring
By following the change in absorbance as a function of pH we will determine the acid dissociation constant or pKa. This technique is not restricted to |
| Core practical 14: Determine the activation energy for the reaction |
|
Fertilizers – Method for determination of free acidity
&basdatefin=&baspays=TZA&baspays2=TZA&basnotifnum=54&basnotifnum2=54&bastypepays=TZA&baskeywords=&project_type_num=1&project_type_id=1&lang_id=EN |
|
Practical Problems Revised Grading Scheme
Calculate the molar absorptivities at 470 and 520 nm of acidic and basic forms of Determination of solution pH by using acid-base indicator (methyl red). |
|
Experiment 6 Titration II – Acid Dissociation Constant
the goal is to determine the equivalence point of the titration. indicate the pH range in which phenolphthalein and methyl red change colors. |
|
DRAGON FRUIT FRESHNESS DETECTOR BASED ON METHYL
Smart label based on methyl red colour indicator can be used to determine the freshness of this non climacteric fruit. This study aimed to develop smart |
| Chapter 9 |
| Synthesis and performance evaluation of adsorbents derived from |
| Q3C (R8) Step 5 - impurities: guideline for residual solvents |
| Statistical optimization for simultaneous removal of methyl red and |
|
PKa of Methyl Red Purpose: The pKa of methyl red - Colby College
of methyl red will be determined by measuring absorbance spectra as a Estimate the expected error in the final result based on the measurement errors |
|
A Simplified Method for Finding the pKa of an Acid–Base Indicator
3 mar 1999 · values are substituted into eq 3 and the pKa is calculated for each set methyl red produces reasonably accurate results (1, 7,8) The pKa |
|
Expt4pdf
The objective of this experiment is to determine the acid dissociation constant for methyl Methyl red is a type of dye: acid o-p-dimethylamino-phenylazo) benzoic With those results, choose the two most suitable wavelengths based on the |
|
Equilibrium and Structural Study of m-Methyl Red in Aqueous
isomers of methyl red that is one of a large number of indicators based upon aminoazo- From the calculated Ka values, the distribution diagram of basic, lowering pH, as a result of the gradual transformation of m-MR from its basic form |
|
A hand book of - For IIT Kanpur
28 juil 2012 · will result in zero marks for the experiment; and a warning against repeating the same “To determine the rate of reaction of A and B using method of D” 3 (a) A procedure (not detailed) that is based on the principles in part 3 The protonated red form (HMR) of methyl red exists in acid solutions When |
|
Exoperiment 5
will be determined and the Ka wil be calculated Methyl red or methyl orange indicator may be used Graph on excel the results for each of the titrations |
|
Practical Problem IChO 2017 - ICho 2019
after one warning will result in being dismissed from the laboratory and zero marks for the Calculate the concentration ratio of basic form and acidic form of methyl red in the can be calculated based on the information given on the label |
|
Exp 13 pH AND ITS MEASUREMENTS Sp07
and when properly chosen can be used to determine the range of the pH of the solution A given indicator is useful for determining pH only in the region in which |
|
Experiment &# 11: Spectroscopic determination of indicator pKa - ULM
the Henderson-Hasselbalch equation will be used to determine pKa values of various indicators Chlorophenol red (33 mg/l) The spectroscopic determination of indicator pKa, involves calculations based on the following 1) What single error would have the greatest effect on the accuracy of the experimental results? |