 Quoi faire avec des résultats de qPCR
Quoi faire avec des résultats de qPCR
2 pouvoir détecter l'amplification fluorescente. Si le Ct a une petite valeur (10-15) le gène est Delta Delta Ct = ?Ct échantillon1 – ?Ct calibrateur.
 Relative quantification
Relative quantification
Two general types of quantification strategies can be performed in qRT-. PCR. The levels of expressed genes 1–2 using ?CP) or 'delta-delta Ct' method.
 Analyzing your QRT-PCR Data The Comparative CT Method (??CT
Analyzing your QRT-PCR Data The Comparative CT Method (??CT
The Comparative CT Method (??CT Method): Data Analysis Example 2 = 0.022 [in excel for s1 simply =STD(select boxes with values of interest)]. ?CT = CT ...
 Understanding qPCR results
Understanding qPCR results
If there is a difference of 2 cycles between two reactions (see figure) we The standard deviation is calculated by the software with the delta Ct value ...
 RT² Profiler PCR Array Gene Expression Analysis Report
RT² Profiler PCR Array Gene Expression Analysis Report
28 mai 2022 Fold Change is then calculated using 2^ (-delta delta CT) formula. The data analysis web portal also plots scatter plot volcano plot
 Comprendre des résultats de qPCR
Comprendre des résultats de qPCR
Donc s'il y a une différence de 2 cycles entre deux réactions (voir image)
 The in vitro and in vivo potency of CT-P59 against Delta and its
The in vitro and in vivo potency of CT-P59 against Delta and its
23 juil. 2021 Rey Olivier Schwartz
 Information on qPCR results
Information on qPCR results
the expression of a gene between two samples. The standard deviation is calculated by the software with the delta Ct value of the technical triplicates.
 Spread of the Delta variant vaccine effectiveness against PCR
Spread of the Delta variant vaccine effectiveness against PCR
16 juil. 2021 region and a larger viral load at symptom onset (-2.4 Ct) for Delta variant infections. The SARS-CoV-2 variant of concern Delta (clade ...
 SARS-CoV-2 variants of concern and variants under investigation in
SARS-CoV-2 variants of concern and variants under investigation in
6 août 2021 Delta variant accounted for approximately 99% of sequenced and 98% genotyped cases from 25 July to 31 July 2021. • PCR cycle threshold (Ct) ...
 How To Perform The Delta-Delta Ct Method
How To Perform The Delta-Delta Ct Method
Here we describe the 2???C T algorithm implemented in the ddCt package The package is designed for the data analysis of quantitative real–time PCR (qRT–PCR) experiemtns in Bioconductor With the ddCt package one can acquire the relative expression of the target gene in different samples
 Analyzing your QRT
Analyzing your QRT
The Comparative CT Method (??CT Method): Data Analysis Example The following table presents data from an experiment where the expression levels of a target (c- myc ) and an endogenous control (GAPDH) are evaluated
 Searches related to 2 delta ct PDF
Searches related to 2 delta ct PDF
All CT accuracy considerations require knowledge of the CT burden The externalload applied to the secondary of a current transformer is called the Òburden Ó The burden is expressed pr eferably in ter ms of the impedance of the load and its r esistance and r eactance components
TaqMan Probes
TaqMan probes depend on the 5'- nuclease activity of the DNA polymerase used for PCR to hydrolyze an oligonucleotide that is hybridized to the target amplicon. TaqMan probes are oligonucleotides that have a fluorescent reporter dye attached to the 5' end and a quencher moeity coupled to the 3' end. These probes are designed to hybridize to an inter...
Molecular Beacons
Like TaqMan probes, Molecular Beacons also use FRET to detect and quantitate the synthesized PCR product via a fluor coupled to the 5' end and a quench attached to the 3' end of an oligonucleotide substrate. Unlike TaqMan probes, Molecular Beacons are designed to remain intact during the amplification reaction, and must rebind to target in every cy...
Scorpions
With Scorpion probes, sequence-specific priming and PCR product detection is achieved using a single oligonucleotide. The Scorpion probe maintains a stem-loop configuration in the unhybridized state. The fluorophore is attached to the 5' end and is quenched by a moiety coupled to the 3' end. The 3' portion of the stem also contains sequence that is...
SYBR Green
SYBR Green provides the simplest and most economical format for detecting and quantitating PCR products in real-time reactions. SYBR Green binds double-stranded DNA, and upon excitation emits light. Thus, as a PCR product accumulates, fluorescence increases. The advantages of SYBR Green are that it is inexpensive, easy to use, and sensitive. The di...
What is a Delta-Delta CT Method?
This is to essentially normalise the gene of interest to a gene which is not affected by your experiment, hence the housekeeping gene-term. To use the delta-delta Ct method, you require Ct values for your gene of interest and your housekeeping gene for both the treated and untreated samples.
Where is the 2 ct method used?
3 Department of Molecular and Cellular Oncology, The University of Texas MD Anderson Cancer Center, Houston, Texas 77230, USA. Background: The 2 -??CT method has been extensively used as a relative quantification strategy for quantitative real-time polymerase chain reaction (qPCR) data analysis.
Is Delta-Delta CT the same as Pfaffl equation?
Many thanks for your message. The delta-delta Ct method assumes your primer efficiencies between your target gene and housekeeping gene are the same (or roughtly the same). However, what would be even better in your case is to use the Pfaffl equation to account for the slight differences in primer efficiencies.
What if cDNA dilution versus Delta CT is close to zero?
If the plot of cDNA dilution versus delta Ct is close to zero, it implies that the efficiencies of the target and housekeeping genes are very similar. If a housekeeping gene cannot be found whose amplification efficiency is similar to the target, then the standard curve method is preferred.
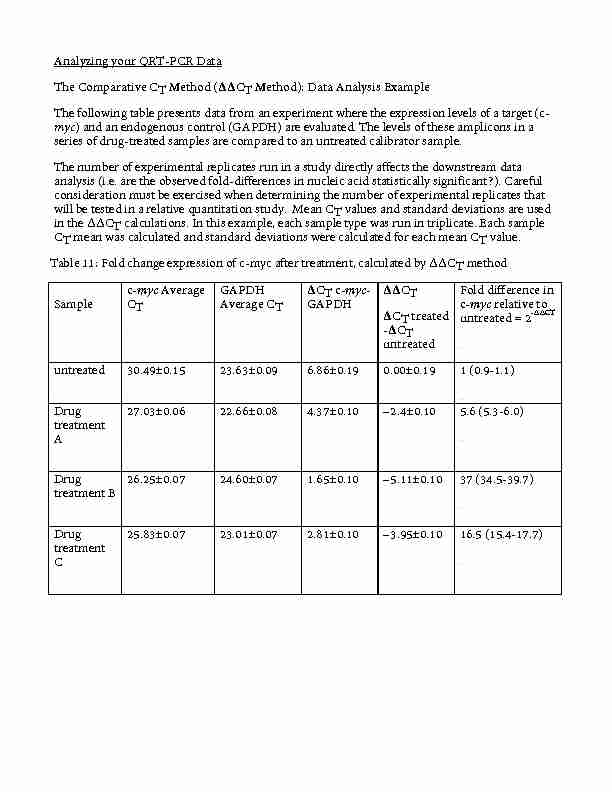
Analyzing your QRT-PCR Data The Comparative CT Method (ΔΔCT Method): Data Analysis Example The following table presents data from an experiment where the expression levels of a target (c-myc) and an endogenous control (GAPDH) are evaluated. The levels of these amplicons in a series of drug-treated samples are compared to an untreated calibrator sample. The number of experimental replicates run in a study directly affects the downstream data analysis (i.e. are the observed fold-differences in nucleic acid statistically significant?). Careful consideration must be exercised when determining the number of experimental replicates that will be tested in a relative quantitation study. Mean CT values and standard deviations are used in the ΔΔCT calculations. In this example, each sample type was run in triplicate. Each sample CT mean was calculated and standard deviations were calculated for each mean CT value. Table 11: Fold change expression of c-myc after treatment, calculated by ΔΔCT method Sample c-myc Average CT GAPDH Average CT ΔCT c-myc- GAPDH ΔΔCT ΔCT treated -ΔCT untreated Fold difference in c-myc relative to untreated = 2-∆∆CT untreated 30.49±0.15 23.63±0.09 6.86±0.19 0.00±0.19 1 (0.9-1.1) Drug treatment A 27.03±0.06 22.66±0.08 4.37±0.10 -2.4±0.10 5.6 (5.3-6.0) Drug treatment B 26.25±0.07 24.60±0.07 1.65±0.10 -5.11±0.10 37 (34.5-39.7) Drug treatment C 25.83±0.07 23.01±0.07 2.81±0.10 -
3.95±0.10 16.5 (15.4-17.7)
Calculate the ΔCT value. Open data up in an excel file: Based on consistency of amplification, choose either 18S or GAPDH as your endogenous control. Calculate the average CT for your endogenous control and each experimental gene as follows: =AVG(select boxes with values of interest) The ΔCT value is calculated by: For example, subtraction of the average GAPDH CT value from the average c-myc CT value of the untreated sample yields a value of 6.86. ΔCT untreated = 30.49 - 23.63 = 6.86 Calculate the standard deviation of CT values and variance of the ΔCT value. The variance of the ΔCT is calculated from the standard deviations of the target and reference values using the formula: s = (s12 + s22)1/2 ; where X1/2 is the square root of X and s= standard deviation. For example, to calculate the standard deviation of the untreated sample ΔCT value: s1 = 0.15 and s12 = 0.022 [in excel for s1 simply =STD(select boxes with values of interest)] ΔCT = CT target - CT reference s2 = 0.09 and s22 = 0.008 s = (0.022 + 0.008)1/2 = 0.17 Therefore, ΔCT untreated = (30.49 ±0.15) - (23.63 ±0.09) = 6.86 ±0.17 Calculate the ΔΔCT value. The ΔΔCT is calculated by: ΔΔCT = ΔCT test sample - ΔCT calibrator sample For example, subtracting the ΔCT of the untreated from the ΔCT of Drug Treatment A yields a value of -2.5. ΔΔCT = 4.37 - 6.86 = -2.5 Calculate the standard deviation of the ΔΔCT value. The calculation of ΔΔCT involves subtraction of the ΔCT calibrator value. This is subtraction of an arbitrary constant, so the standard deviation of the ΔΔCT value is the same as the standard
deviation of the ΔCT value. Therefore, ΔΔCT Drug Treatment A sample = ΔΔCT = 4.37±0.10 - 6.86±0.17 = -2.5±0.10 Standard deviation of the ΔΔCT value is the same as the standard deviation of the ΔCT value Incorporating the standard deviation of the ΔΔCT values into the fold- difference. Fold-differences calculated using the ΔΔCT method are usually expressed as a range, which is a result of incorporating the standard deviation of the ΔΔCT value into the fold- difference calculation. The range for targetN, relative to a calibrator sample, is calculated by: 2-ΔΔCt with ΔΔCT + s and ΔΔCT - s, where s is the standard deviation of the ΔΔCT value. For example, the drug-treatment A sample has a 5.3 to 6.0-fold difference in expression of the targetN relative to the untreated (calibrator) as indicated below. ΔΔCT +s=-2.5+0.1=-2.4 2-ΔΔCt = 2- (-2.4) = 5.3 and ΔΔCT +s=-2.5-0.1=-2.6 2-ΔΔCt = 2- (-2.6) = 6.0 At this point to get the true fold change, we take the log base 2 of this value to even out the scales of up regulated and down regulated genes. Otherwise upregulated has a scale of 1-infinity while down regulated has a scale of 0-1. Once you have your fold changes, you can then look into the genes that seem the most interesting based on this data. There are hundreds of resources online that will tell you what the gene does, what pathways it is involved in, etc. We will start by going to the website created by the Barres Lab team from Stanford that wrote the RNA Seq paper on the various isolated cells in the NS. Find the link below: Works better in Safari http://web.stanford.edu/group/barres_lab/brain_rnaseq.html
QRT-PCR - Analyzing your Data - Further Notes for consideration and questions for discussion. Based on your amplification plots, the computer will determine the best threshold to set whereby themost amplification plots are in a linear growth phase. Once the threshold is set, the cycle at which each
amplification curve crossed that threshold is determined and assigned as the CT for that sample. With
this data, you will work in groups and proceed to calculate the change in expression values for each gene
in liver and brain tissue. The CT data is used to determine the amount of each gene/mRNA present relative to each sample. The table below shows the average CT results for the expression of VEGF in healing Achilles tendons in mice immediately post-op and 1 day post-op, and how these CTs are manipulated to determine ΔCT, ΔΔCT, and the relative amount of VEGF mRNA in terms of fold change. ∆CT is calculated bysubtracting the CT for VEGF for the sample from the CT for the endogenous control (in this case 18S).
The calculation of ΔΔCT involves subtraction by the ΔCT reference sample value (in this case from the
wild type for one calculation and from day 0 for a second calculation). The range given for VEGF in wild type mice relative mutant mice is determined by evaluating the expression: 2 -ΔΔCTData can be graphed in a variety of ways, once expression has been determined, for easier visualization.
Below are examples of how the data in the table may look. Only the Day0 and Day1 points are shown in the table while the graphs show the data through Day7 post-op. The scatter plot displays the difference in expression of VEGF in both the wild type and mutant mice using the Day 0 data for eachmouse type as the reference. Notice that overall expression decreases for both mice types as healing
progresses, though the decrease is greater for the mutants. The bar graph shows the difference in expression of VEGF in the wild type and mutant mice at each day post-op using the wild type for thatday as the reference. Notice that the wild type mice have the lower expression on each day except day 2
and day 7, when their expression is higher than the mutants.Once calculations are done, you can further investigate the genes that you are still interested in by going
online and finding databases that help you determine gene function and rolls in pathways. There are many tools available free online - Gene Expression Omnibus (GEO), Online Mendelian Inheritance inMan (OMIM), and Biocyc, just to name a few. We will investigate this a little bit together if time today
and finish up on the last day.Questions for Discussion
1. Which genes were most were more highly expressed in the brain?
2. Which genes were more highly expressed in the liver?
3. Based on the functions of these genes, does it make sense that they are differentially expressed in
these two organs? Use two of the genes to help explain why or why not.quotesdbs_dbs28.pdfusesText_34[PDF] delta delta ct calculation
[PDF] comment faire un transect
[PDF] comment réaliser un transect de végétation
[PDF] exemple de transect
[PDF] comment réaliser un transect végétal
[PDF] transect botanique
[PDF] transect definition
[PDF] protocole pcr taqman
[PDF] analyse résultats pcr quantitative
[PDF] pcr protocole pdf
[PDF] qpcr sybr green principe
[PDF] protocole rt pcr
[PDF] quiz sur lespace facile
[PDF] pcr en temps réel protocole
